Data processing and visualisation
Overview
Teaching: 20 min
Exercises: 40 minQuestions
How can I create a quick plot of my CMIP data?
Objectives
Import the xarray library and use the functions it contains.
Convert precipitation units to mm/day.
Calculate and plot the precipitation climatology.
Use the cmocean library to find colormaps designed for ocean science.
As a first step towards making a visual comparison of the ACCESS-CM2 and ACCESS-ESM1-5 historical precipitation climatology, we are going to create a quick plot of the ACCESS-CM2 data.
accesscm2_pr_file = 'data/pr_Amon_ACCESS-CM2_historical_r1i1p1f1_gn_201001-201412.nc'
We will need a number of the libraries introduced in the previous lesson.
import xarray as xr
import cartopy.crs as ccrs
import matplotlib.pyplot as plt
import numpy as np
Since geographic data files can often be very large, when we first open our data file in xarray it simply loads the metadata associated with the file (this is known as “lazy loading”). We can then view summary information about the contents of the file before deciding whether we’d like to load some or all of the data into memory.
dset = xr.open_dataset(accesscm2_pr_file)
print(dset)
<xarray.Dataset>
Dimensions: (bnds: 2, lat: 144, lon: 192, time: 60)
Coordinates:
* time (time) datetime64[ns] 2010-01-16T12:00:00 ... 2014-12-16T12:00:00
* lon (lon) float64 0.9375 2.812 4.688 6.562 ... 355.3 357.2 359.1
* lat (lat) float64 -89.38 -88.12 -86.88 -85.62 ... 86.88 88.12 89.38
Dimensions without coordinates: bnds
Data variables:
time_bnds (time, bnds) datetime64[ns] ...
lon_bnds (lon, bnds) float64 ...
lat_bnds (lat, bnds) float64 ...
pr (time, lat, lon) float32 ...
Attributes:
CDI: Climate Data Interface version 1.9.8 (https://mpi...
source: ACCESS-CM2 (2019): \naerosol: UKCA-GLOMAP-mode\na...
institution: CSIRO (Commonwealth Scientific and Industrial Res...
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: standard
branch_time_in_child: 0.0
branch_time_in_parent: 0.0
creation_date: 2019-11-08T08:26:37Z
data_specs_version: 01.00.30
experiment: all-forcing simulation of the recent past
experiment_id: historical
external_variables: areacella
forcing_index: 1
frequency: mon
further_info_url: https://furtherinfo.es-doc.org/CMIP6.CSIRO-ARCCSS...
grid: native atmosphere N96 grid (144x192 latxlon)
grid_label: gn
initialization_index: 1
institution_id: CSIRO-ARCCSS
mip_era: CMIP6
nominal_resolution: 250 km
notes: Exp: CM2-historical; Local ID: bj594; Variable: p...
parent_activity_id: CMIP
parent_experiment_id: piControl
parent_mip_era: CMIP6
parent_source_id: ACCESS-CM2
parent_time_units: days since 0950-01-01
parent_variant_label: r1i1p1f1
physics_index: 1
product: model-output
realization_index: 1
realm: atmos
run_variant: forcing: GHG, Oz, SA, Sl, Vl, BC, OC, (GHG = CO2,...
source_id: ACCESS-CM2
source_type: AOGCM
sub_experiment: none
sub_experiment_id: none
table_id: Amon
table_info: Creation Date:(30 April 2019) MD5:e14f55f257cceaf...
title: ACCESS-CM2 output prepared for CMIP6
variable_id: pr
variant_label: r1i1p1f1
version: v20191108
cmor_version: 3.4.0
tracking_id: hdl:21.14100/b4dd0f13-6073-4d10-b4e6-7d7a4401e37d
license: CMIP6 model data produced by CSIRO is licensed un...
CDO: Climate Data Operators version 1.9.8 (https://mpi...
history: Tue Jan 12 14:50:25 2021: ncatted -O -a history,p...
NCO: netCDF Operators version 4.9.2 (Homepage = http:/...
We can see that our dset object is an xarray.Dataset,
which when printed shows all the metadata associated with our netCDF data file.
In this case, we are interested in the precipitation variable contained within that xarray Dataset:
print(dset['pr'])
<xarray.DataArray 'pr' (time: 60, lat: 144, lon: 192)>
[1658880 values with dtype=float32]
Coordinates:
* time (time) datetime64[ns] 2010-01-16T12:00:00 ... 2014-12-16T12:00:00
* lon (lon) float64 0.9375 2.812 4.688 6.562 ... 353.4 355.3 357.2 359.1
* lat (lat) float64 -89.38 -88.12 -86.88 -85.62 ... 86.88 88.12 89.38
Attributes:
standard_name: precipitation_flux
long_name: Precipitation
units: kg m-2 s-1
comment: includes both liquid and solid phases
cell_methods: area: time: mean
cell_measures: area: areacella
We can actually use either the dset['pr'] or dset.pr syntax to access the precipitation
xarray.DataArray.
To calculate the precipitation climatology, we can make use of the fact that xarray DataArrays have built in functionality for averaging over their dimensions.
clim = dset['pr'].mean('time', keep_attrs=True)
print(clim)
<xarray.DataArray 'pr' (lat: 144, lon: 192)>
array([[1.8461452e-06, 1.9054805e-06, 1.9228980e-06, ..., 1.9869783e-06,
2.0026005e-06, 1.9683730e-06],
[1.9064508e-06, 1.9021350e-06, 1.8931637e-06, ..., 1.9433096e-06,
1.9182237e-06, 1.9072245e-06],
[2.1003202e-06, 2.0477617e-06, 2.0348527e-06, ..., 2.2391034e-06,
2.1970161e-06, 2.1641599e-06],
...,
[7.5109556e-06, 7.4777777e-06, 7.4689174e-06, ..., 7.3359679e-06,
7.3987890e-06, 7.3978440e-06],
[7.1837171e-06, 7.1722038e-06, 7.1926393e-06, ..., 7.1552149e-06,
7.1576678e-06, 7.1592167e-06],
[7.0353467e-06, 7.0403985e-06, 7.0326828e-06, ..., 7.0392648e-06,
7.0387587e-06, 7.0304386e-06]], dtype=float32)
Coordinates:
* lon (lon) float64 0.9375 2.812 4.688 6.562 ... 353.4 355.3 357.2 359.1
* lat (lat) float64 -89.38 -88.12 -86.88 -85.62 ... 86.88 88.12 89.38
Attributes:
standard_name: precipitation_flux
long_name: Precipitation
units: kg m-2 s-1
comment: includes both liquid and solid phases
cell_methods: area: time: mean
cell_measures: area: areacella
Now that we’ve calculated the climatology, we want to convert the units from kg m-2 s-1 to something that we are a little more familiar with like mm day-1.
To do this, consider that 1 kg of rain water spread over 1 m2 of surface is 1 mm in thickness and that there are 86400 seconds in one day. Therefore, 1 kg m-2 s-1 = 86400 mm day-1.
The data associated with our xarray DataArray is simply a numpy array,
type(clim.data)
numpy.ndarray
so we can go ahead and multiply that array by 86400 and update the units attribute accordingly:
clim.data = clim.data * 86400
clim.attrs['units'] = 'mm/day'
print(clim)
<xarray.DataArray 'pr' (lat: 144, lon: 192)>
array([[0.15950695, 0.16463352, 0.16613839, ..., 0.17167493, 0.17302468,
0.17006743],
[0.16471735, 0.16434446, 0.16356934, ..., 0.16790195, 0.16573453,
0.1647842 ],
[0.18146767, 0.17692661, 0.17581128, ..., 0.19345854, 0.18982219,
0.18698342],
...,
[0.64894656, 0.64607999, 0.64531446, ..., 0.63382763, 0.63925537,
0.63917372],
[0.62067316, 0.61967841, 0.62144403, ..., 0.61821057, 0.6184225 ,
0.61855632],
[0.60785395, 0.60829043, 0.60762379, ..., 0.60819248, 0.60814875,
0.6074299 ]])
Coordinates:
* lon (lon) float64 0.9375 2.812 4.688 6.562 ... 353.4 355.3 357.2 359.1
* lat (lat) float64 -89.38 -88.12 -86.88 -85.62 ... 86.88 88.12 89.38
Attributes:
standard_name: precipitation_flux
long_name: Precipitation
units: mm/day
comment: includes both liquid and solid phases
cell_methods: area: time: mean
cell_measures: area: areacella
We could now go ahead and plot our climatology using matplotlib,
but it would take many lines of code to extract all the latitude and longitude information
and to setup all the plot characteristics.
Recognising this burden,
the xarray developers have built on top of matplotlib.pyplot to make the visualisation
of xarray DataArrays much easier.
fig = plt.figure(figsize=[12,5])
ax = fig.add_subplot(111, projection=ccrs.PlateCarree(central_longitude=180))
clim.plot.contourf(ax=ax,
levels=np.arange(0, 13.5, 1.5),
extend='max',
transform=ccrs.PlateCarree(),
cbar_kwargs={'label': clim.units})
ax.coastlines()
plt.show()
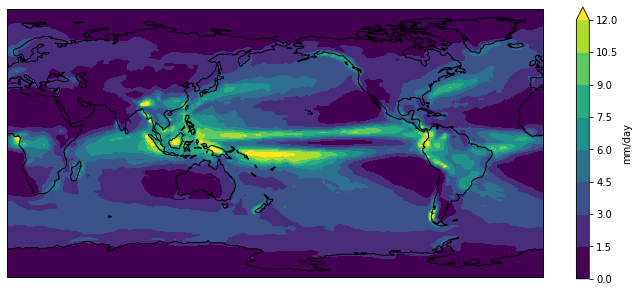
The default colorbar used by matplotlib is viridis.
It used to be jet,
but that was changed a couple of years ago in response to the
#endtherainbow campaign.
Putting all the code together (and reversing viridis so that wet is purple and dry is yellow)…
import xarray as xr
import cartopy.crs as ccrs
import matplotlib.pyplot as plt
import numpy as np
accesscm2_pr_file = 'data/pr_Amon_ACCESS-CM2_historical_r1i1p1f1_gn_201001-201412.nc'
dset = xr.open_dataset(accesscm2_pr_file)
clim = dset['pr'].mean('time', keep_attrs=True)
clim.data = clim.data * 86400
clim.attrs['units'] = 'mm/day'
fig = plt.figure(figsize=[12,5])
ax = fig.add_subplot(111, projection=ccrs.PlateCarree(central_longitude=180))
clim.plot.contourf(ax=ax,
levels=np.arange(0, 13.5, 1.5),
extend='max',
transform=ccrs.PlateCarree(),
cbar_kwargs={'label': clim.units},
cmap='viridis_r')
ax.coastlines()
plt.show()
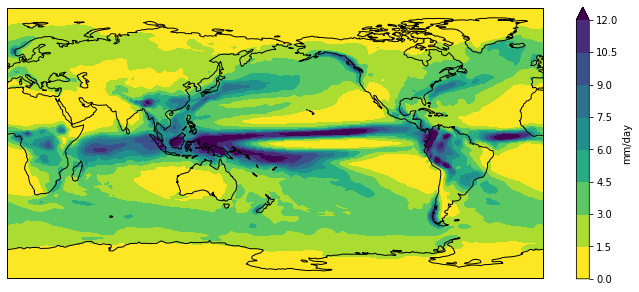
Color palette
Copy and paste the final slab of code above into your own Jupyter notebook.
The viridis color palette doesn’t seem quite right for rainfall. Change it to the “haline” cmocean palette used for ocean salinity data.
Solution
import cmocean ... clim.plot.contourf(ax=ax, ... cmap=cmocean.cm.haline_r)
Season selection
Rather than plot the annual climatology, edit the code so that it plots the June-August (JJA) season.
(Hint: the groupby functionality can be used to group all the data into seasons prior to averaging over the time axis)
Solution
clim = dset['pr'].groupby('time.season').mean('time', keep_attrs=True) clim.sel(season='JJA').plot.contourf(ax=ax,
Add a title
Add a title to the plot which gives the name of the model (taken from the
dsetattributes) followed by the words “precipitation climatology (JJA)”Solution
model = dset.attrs['source_id'] title = f'{model} precipitation climatology (JJA)' plt.title(title)
Key Points
Libraries such as xarray can make loading, processing and visualising netCDF data much easier.
The cmocean library contains colormaps custom made for the ocean sciences.